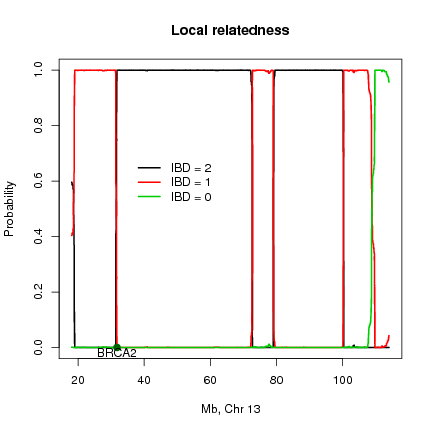
This method estimates the probability of sharing alleles identity by descent (IBD) across the genome and can also be used for mapping disease loci using distantly related individuals
The method is implemented in an R package and as a commandline based C++ program embeded in the R package. The R code can be used to find and visualize the tracts of relatedness between a pair of individuals. The commandline version has under 20% of the running time when running all pairs compared to a single pair, it however has the the same speed for running a single pair analysis. For analysis linkage only the C++ version is implemented.
If you have the devtools packages (https://github.com/hadley/devtools) installed in R then you can install the package i R directly from github
library(remote)
remotes::install_github("aalbrechtsen/relate")
#or using devtools
library(devtools)
install_github("aalbrechtsen/relate")
download the code
git clone https://github.com/aalbrechtsen/relate.git
go to the scr folder that contains the C++ files type
./install.sh
If you do not have the devtools package (and dont want to install it) then you will have to build the R package
first download the code (you need to have a clean version without the compiled c++ code)
git clone https://github.com/aalbrechtsen/relate.git
R CMD build relate
or build and install
R CMD build relate
R CMD INSTALL Relate_<add version number>.tar.gz
- The manual can be found on the wiki [http://www.popgen.dk/software/index.php/Relate]
library(Relate)
example(relate)
./src/relateHMM
#standard input format, single individuals
./src/relateHMM -o data/options.single.pair.txt -g data/dat_small.geno -p data/dat_small.pos -c data/dat_small.chr
#standard input format, multiple individuals
./src/relateHMM -o data/options.all.pair.txt -g data/dat_small.geno -p data/dat_small.pos -c data/dat_small.chr -d data/dat_small.keep
#plink input format
./src/relateHMM -o data/options.single.pair.txt -plink data/500